Frequently Asked Questions
Cancer, ageing, stem cells and cloning are just some of the many evokative words that have been associated with epigenetic research. What is epigenetics? What are stem cells? What impact do these things have on ordinary lives? These important FAQs among others are addressed here.
- What is a genome?
- What is a chromosome?
- What is epigenetics?
- What is the relationship between genetics and epigenetics?
- What is a nucleosome?
- How can nucleotides and proteins be modified into epigenetic tags?
- How does an epigenetic tag work?
- Which experimental technologies and approaches are applied in epigenetic research?
- Which organisms are used in epigenetic research?
- How were chromosomes discovered?
- What is chromatin, euchromatin and heterochromatin?
- When did epigenetic traits originate in evolution?
- What is meant with the term 'histone code'?
- What is epigenetic imprinting?
- Is eugenics related to epigenetics?
- What do stem cells do?
- What can we use stem cells for?
- What is nuclear transfer?
- What's the difference between embryonic and adult stem cells?
- What is the difference between epigenetics and epigenomics?
1. What is a genome?
The genome represents the entire genetic information stored in the chromosomes of an organism. It enables the fertilized egg (i.e. the zygote) or an embryonic stem cell to complete development into an adult organism containing all organs, physiological functions and structures necessary for living and reproduction. This genetic information is maintained in the nucleotide sequence of DNA, where DNA is an integrated part of the chromosomes. Molecular DNA duplication-processes guarantee the genome to be present in nearly all cells of a respective organism. The genome also serves as a sanctuary for long term storage of the genetic information established over millions of years of evolution. To this end, replica copies of each DNA strand are produced by duplication, which, if damaged through mutation, serve as blueprints for each other to correct occurring errors.
2. What is a chromosome?
A chromosome is a large macromolecule which contains part of the genetic information stored in the genome. The core of the chromosome-molecule is a double stranded DNA string which is interacting non-covalently with a large number of proteins (chromatin proteins) and shorter chains of ribonucleic acid (RNA). The DNA of a chromosome within a human cell spans several centimeters and has to be compacted in order to fit into an average cell’s nucleus of 7 micrometer diameter. This compaction is mediated by the proteins associated with the DNA. Each chromosome must contain at least one origin of replication. While prokaryotic microorganisms usually contain a circular chromosome, the ends of linear chromosomes of eurkaryotes must be protected against degradation by special proteins. The ends of linear chromosomes are called telomeres.
3. What is epigenetics?
In 1942 Conrad Hal Waddington defined epigenetics as 'the branch of biology which studies the causal interactions between genes and their products which bring the phenotype into being'. In modern sense the term 'epigenetics' describes heritable changes in genome function that occur without a change in nucleotide sequence within the DNA. For example, when a cell established a particular pattern of 'active' and 'non-active' genes this same pattern will be passed on to a daughter cell even though during cell division all genes are 'shut off' and chromosomes have become tightly wrapped up or condensed. This process allows development of different structures and organs during development. The nucleotides within DNA sometimes become chemically modified. A number of combined, nearby modifications may represent a particular pattern. Such a pattern may serve as a template for passing on its informative message in the form of specific chemical modifications to other molecules. Particular aminoacid groups (e.g. lysines) within proteins such as histones may be modified through acetylation or methylation and serve as transmitters of such information. Such chemical modification patterns are called epigenetic tags. For more information www.epigenome-noe.net/aboutus/epigenetics.php
4. What is the relationship between genetics and epigenetics?
The term 'Genetics' was coined by William Bateson in 1905. However, the term 'gene' is based on ideas of Charles Darwin and Hugo de Vries: In 1878 Hugo de Vries visited Darwin, who directly stimulated de Vries to shift from physiological to evolutionary and genetic studies. The source of de Vries‘ inspiration was Darwin‘s speculation on heredity, i.e. the provisional hypothesis of pangenesis. According to pangenesis, hereditary characters are part of tiny cellular particles called gemmules. All cells produce gemmules during develoment, growth and later life. Gemmules then migrate from somatic to germ cells, where they collect to pass inherited characters to the next generation. This, actually was Darwin‘s famous lamarckist statement! Hugo de Vries suggested a fundamental (and correct) modification: he abandoned Darwin‘s key notion of the migration of gemmules across cell boundaries, and rechristened gemmulae as 'pangenes'. Wilhelm Ludvig Johannsen in 1909 finally coined the term 'gene' (for further reeding see (1)). Only later Hermann Muller's discovery in 1927, that genes can be altered by gamma radiation and are therefore real physical entities, and Watson's and Crick's discovery of the DNA structure in 1953 led to genes as the major focus of understanding the morphology of phenotypes, life histories and physiology. In 1942, Waddington defined epigenetics as 'the branch of biology which studies the causal interactions between genes and their products which bring the phenotype into being.' Because this objective was always the driving force of biology since Darwin and before, we now understand from a traditional, historical perspective that there is a tight, inseparable kinship between genetics and epigenetics. Today, epigenetics can be best envisaged as the causal, logical and consequential modern successor of genetics.
1. Gould, S. J. 2002. The structure of evolutionary theory. Belknap Press of Harvard University Press, Cambridge, Mass
5. What is a nucleosome?
The DNA molecules of eukaryotes are linear chains of nucleotides with lengths in the centimeter range. The size of the nuclei in which this DNA resides is on average only 7 micrometer in diameter. The evolution of such long molecules was possible only because the DNA strings were tightly folded around proteins leading to the compaction of a chromosome. The major protein complex with DNA wrapped around itself is called a nucleosome. Each nucleosome consists of four proteins called histones. Nucleosomes are positively charged at the N-termini of their histones. There are two copies of each histone molecule for every 200 bp of DNA. The histones are called H2A, H2B, H3 and H4. Another, larger single histone molecule, H1 is called a linker histone. It binds to DNA molecules which cross over each other and seals the complex at the exterior of the nucleosome such that the DNA does not unfold. The DNA of a typical eukaryotic genome is about 1m long. The compaction of DNA into nucleosomal chains would reduce this length to about 15 to 20 cm. In a 7 micrometer nucleus this still requires further condensation. Therefore, nucleosomes cross-link to each other and additional proteins such as HMG14 and HMG1. Like H1, HMG proteins have a preference for binding crossed DNA molecules. They are also major linker DNA-binding proteins and help in conjunction with the nucleosomes to form higher order chromsome structures.
6. How can nucleotides and proteins be modified into epigenetic tags?
DNA frequently becomes methylated by an enzyme called DNA methyltransferase. DNA methylation is a type of chemical modification which involves the addition of a methyl group to the carbon-5 of the cytosine pyrimidine ring. This methyl group can be sensed by proteins which then themselves modify other proteins posttranslationally. The pattern of posttranslational change creates changes in electric charge within chromatin and this leads to different degrees of chromatin condensation. It also determines whether genes can be transcribed or become silent within highly compacted chromatin. Silence of genes means, that their encoded genetic information is not transribed into mRNA and subsequently it will not be translated into protein molecules. There are many ways of how electric charges between nucleotide molecules and proteins can be altered. These variances of charge are identical with what people call epigenetic tags.
In addition to DNA methylation (i.e. a form of alkylation), proteins can become posttranslationally modified through: methylation (histone methyltransferase), phosphorylation (kinases), acetylation, isoprenylation, glycosylation, ubiquitination, SUMOylation, poly(ADP)ribosylation (PARP).
7. How does an epigenetic tag work?
While different models are discussed, the question is best provisionally explained by a classical, simplified example(1,2): The phosphate groups connecting the nucleotides of a DNA chain are negatively charged. In naked dsDNA they would repel each other, and the DNA would represent an 'open chromatin' conformation. However, the extruding N termini of nucleosomes are positively charged. Thus, in chromatin consisting only of DNA and nucleosomes the positive histone N-termini would interact with the negative phosphate groups such that the chromatin is highly compacted ('closed chromatin'). There are high levels of H1 linker histones in this chromatin. In a closed chromatin environment genes cannot be transcribed as the transcription factors are sterically hindered to trigger mRNA synthesis - the genes are 'silenced'. The condensed chromatin however can be relaxed by covalently linking acetylgroups (CH3COO-) especially to the amino groups of Lys of the core histones H3 and H4. Acetylation brings in a negative charge and neutralizes the interaction of the N termini with the phosphate groups. As a consequence, the condensed chromatin is transformed into a transiently relaxed structure (see Fig.1) which allows genes to be transcribed. The enzyme catalyzing acetylation is called histone acetyltransferase. Naively one might assume that starting from a zygote, an organism should successively activate all available genes during development in order to live. Thus, at adult age, all genes should be active. However, the simultaneous activity of all genes would produce an uncontrolable chaos of gene expression patterns not allowing coordinated cell- and organ-differentiation. Therefore, many genes need to be more or less permanently inactivated after they have done their job. Such a status can be triggered and maintained by an epigenetic tag. In our example the tag is the methylation of cytosine. Genes which are methylated by a DNA methyltransferase are recognized by the protein MeCP2 which binds to the methylated nucleotides. This protein is complexed with histone deacetylase. Once MeCP2 binds to methylated DNA, histone deacetylase removes the acetyl groups, and the chromatin becomes condensed and inaccessable again for transcription factors. The silenced chromatin can be maintained over most of an organisms lifespan. An example for a protein mediating such a task is encoded by the polycomb gene.
1. Jones, P. L., G. J. Veenstra, P. A. Wade, D. Vermaak, S. U. Kass, N. Landsberger, J. Strouboulis, and A. P. Wolffe. 1998. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet 19:187-91.
2. Nan, X., H. H. Ng, C. A. Johnson, C. D. Laherty, B. M. Turner, R. N. Eisenman, and A. Bird. 1998. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393:386-9.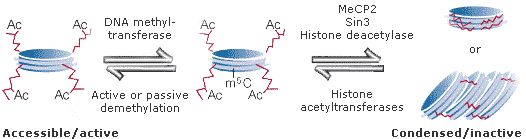
8. Which experimental technologies and approaches are applied in epigenetic research?
Structural molecular biology and biochemistry
Cell biology
Developmental biology
Classical genetics
Advanced imaging
Genomics
Proteomics
Bioinformatics
Mathematical analysis
9. Which organisms are used in epigenetic research?
Epigenetics is studied in different organisms. They represent all kingdoms of life, i.e. eubacteria, archaea, fungi, animals and plants. It frequently includes comparative analysis from an evolutionary perspective. Examples of some well understood research model organisms are: Escherichia coli, Saccharomyces cerevisiae, Arabidopsis thaliana, Hydra hydra, Caenorhabditis elegans, Drosophila melanogaster, Danio rerio, Xenopus laevis, Mus musculus. Analysis of some human diseases also help to advance an understanding of epigenetic mechanisms and the underlying causes of certain diseases.
10. How were chromosomes discovered?
The discovery of the chromosome was descriptive from the beginning and inseparably interwoven with the discoveries of the cell and the nucleus. All findings became possible only after Leeuwenhoek´s invention of the microscope in 1674. In 1831 Robert Brown described the 'areola' in orchids being constantly detectable in all cells. He called this areola'the nucleus of the cel'.1838 M. J. Schleiden´s incorrect epigenetic theory claimed that a cell-nucleus is created de novo from the fluid of the cell. This served as a classical antithesis to Edouard van Beneden´s 1883 discovery that chromosomes are individual entities. In 1842 Karl Wilhelm von Nägeli discovered subcellular structures that would later became known as chromosomes. He had observed the 'idioplasma', a network of string like bodies which he falsely assumed to form an interlinked network throughout the entire organism. In 1873 Schneider had described the indirect division of the nucleus with a 'Kernfigur'(nuclear figure) and an 'achromatic spindle'. In 1883, Edouard van Beneden found that after fertilization of the germ cells of the nematode Ascaris megalocephala the chromosomes of the male nucleus do not fuse with those of the oocyte nucleus. Therefore, they are distinct entities.This was the empirical foundation of Mendel´s rules, but their connection was found only several years later. Van Beneden did not yet use the word chromsome, which was later coined by Waldyer in 1888. The term reflected the staining behavior of chromosomes after using specific dyes.
11. What is chromatin, euchromatin and heterochromatin?
Chromatin is the molecular substance of a chromosome. It consists of a complex of DNA, RNA and protein in eukaryotic cells. Frequently, people encounter pictures of chromosomes which have a striped pattern of stronger and lighter staining. What are these chromosome bands? Do they represent genes? No, they do not necessarily represent genes nor do they automatically correspond to stretches lacking genes. Chromosomes often presented to the public are most highly compacted metaphase chromosomes or the giant chromosomes (i.e. polytene chromosome) of Drosophila larvae from salivary gland cells. The latter are made up of about 2000 DNA double strands arranged parallel to each other. A single band of a Drosophila giant chromosome can contain about 50.000 nucleotides in a row. But what makes its staining different to an adjacent band? What we see as dark bands is a consequence of an increased concentration of DNA within variably, compacted chromatin that differs in staining on average every 50 kb. But there is also a gradient of diverse higher order degrees of compaction. Further, in the life of a chromosome it alters its appearance. During the so-called interphase it is relaxed with a lot of open chromatin (see FAQ 7). During this phase, some entire chromosomes but mostly only parts thereof retain a strong staining. These subnuclear features are known since the early days of cytology. In 1928, Heitz introducd for them the term 'heterochromatin'. Euchromatin, on the other hand, is highly decondensed chromatin. Chromosomal regions in the genome which lack high numbers of genes are normally compacted in heterochromatin while chromosomal regions with high concentrations of transcribed genes are part of relaxed euchromatin. But these patterns change during development depending on the pattern of particular epigenetic tags present in the chromatin (see FAQ 8).
12. When did epigenetic traits originate in evolution?
It is now believed that the first coding nucleotide-chains on earth were RNA molecules. They represented the first replicators in an RNA world roughly 3.5 billion years ago. There are five arguments for an initial RNA world: 1. Prebiotic synthesis of ribose is simpler than synthesis of DNA. 2. The nucleophily of 2´,3´OH groups is much higher than for 3´OH hydroxyls of deoxyribose increasing reactive flexibility. 3. RNA chains are chemically more stable than DNA as their basepair interactions are stronger. Therefore, RNA is capable of forming an enormous spectrum of secondary structures with the potential to evolve catalytic function. 4. DNA replication starts with RNA synthesis in all organisms. 5. Ribonucleotides are the universal precursors in cellular biosynthesis of deoxyribonucleotides (DNA). On the early earth, the transition from an RNA- to a DNA-world did not affect the nucleotide sequence. Therefore, this transition likely represents the first epigenetic modification in early evolution. If peptides, such as chaperones, accompanied early replication is not known, but sophisticated proteins may not have been obligatory as RNA is self-catalytic. Thus, epigenetic modification of nucleotide chains may have been an intrinsic property of life right at the very beginning of organismic evolution.
13. What is meant with the term 'histone code'?
Histone proteins can be modified as epigenetic tags by modifying enzymes in a substrate-specific manner. Prominent are methylation, acetylation and phosphorylation of specific Lysines, Arginines and Serines within the aminoacid sequences of the tails of histones. Imprinting, or silencing, is the suppression of certain genes on chromosomes, depending on from which parent they were received. Combinations of these chemical alterations are thought to modify the structure and function of chromatin. The particular combination of these epigenetic tags may represent various types of chromatin and are thought to represent a histone code in analogy to the genetic code.
14. What is epigenetic imprinting?
The molecular mechanisms responsible for imprinting are defined by the inheritance of epigenetic tags (see FAQ 7) from cell generation to cell generation and from the parents to their offspring. Classically, epigenetic imprinting, or silencing, is the suppression of certain genes on chromosomes, depending on from which parent they were received. When DNA is passed to daughter cells after fertilization of an egg by a sperm, certain alleles can become active only if they were received from the mother, others only if they came from the father. If a gene is suppressed through imprinting from one parent, and the allele from the other parent is not expressed because of mutation, neither can act and the child will be deficient. Genetic imprinting has also been defined as the gamete-of-origin dependent modification of phenotype.
15. Is eugenics related to epigenetics?
Eugenics and epigenetics are not interrelated. The term eugenics is derived from Greek and means 'well born' or 'good breeding'. Its concept was first formulated by Charles Darwin´s cousin Sir Francis Galton in 1865, although he didn't use the term eugenics until 1883. Eugenics is a social philosophy which advocates the supposed improvement of human hereditary qualities. Proposed means of doing so have included (but are not limited to) birth control, selective breeding, genetic engineering, racial hygiene, and even extermination.
16. What do stem cells do?
When you cut yourself, your body's defences are rapidly initiated at the wound site. Your own natural stem cells are recruited to make new skin. Different kinds of stem cells are present in each of your body organs to repair damaged tissue. These are adult stem cells. Scientists are excited about harnessing the power within these special cells to create more organic ways of treating different diseases. Reinforcing the body's own therapeutic strategies has many advantages over traditional chemical treatments, for example chemotherapy, which can be devastating for patients.
17. What can we use stem cells for?
Patients' own adult stem cells have been successfully used to treat a number of conditions, like Parkinson's disease and multiple sclerosis. If you take adult stem cells from other people, they might not be genetically compatible with the patient. Transplanted adult stem cells can be rejected. Another way to avoid rejection of stem cell transplants is to use a much more controversial method. The way that Dolly the Sheep was created. Scientists can take the nucleus out of a fertilised egg cell, and replace this with a nucleus from the patient. This reprogrammed egg cell can be used to clone more cells with the adult nucleus. The resultant cell line could then be used to treat the patient. Obviously not everyone feels comfortable about this technology, because it involves using embryonic tissue.
18. What is nuclear transfer?
Scientists can take the nucleus out of a fertilised egg cell, and replace this with a nucleus from the patient. This reprogrammed egg cell can be used to clone more cells with the adult nucleus. The resultant cell line could then be used to treat the patient. Obviously not everyone feels comfortable about this technology, because it involves using embryonic tissue.
19. What's the difference between embryonic and adult stem cells?
So what's the advantage of using stem cells from the embryo that have been reprogrammed in this way? The main advantage is that you can derive a greater variety of cell types. Embryonic stem cells, as they are called, have the potential to be any body cell. Adult stem cells on the other hand have partly committed to different cell fates. So, for example, you can only make muscle cells from adult stem cells found in muscle. The disadvantage to using embryonic stem cells, is that they have the potential to become cancerous. This has not been observed with adult stem cells. When scientists understand how these cells are programmed to become different cell types, embryonic stem cells could be used in therapies. They can also be used to test drugs, which means that animals don't necessarily need to be used.
20. What is the difference between epigenetics and epigenomics?
In eukaryotic cells, the DNA is packaged within the nucleus in a structure called chromatin. This structure is a highly dynamic nucleoprotein complex that plays a central role in regulating how and when DNA is copied and transcribed into RNA. Thus chromatin states vary from cell type to cell type and along chromosomes. The epigenome refers to these states at the whole genome level. Typically, a multi-cellurar organism will be characterized by one genome, but by as many epigenomes as there are cell types.
Epigenetics encompasses all processes that lead to heritable changes in gene expression (during development or across generations) without changes in the DNA sequence itself. In eukaryotes, chromatin is at the heart of most epigenetic processes. Thus, the study of epigenetics often crosses that of epigenomes (epigenomics).


